GATK Somatic SNV+Indel+CNV+SV
最近准备为sliverworkspace 图形化生信平台开发报告设计器,需要一个较为复杂的pipeline作为测试数据,就想起来把之前的 [[满分室间质评之GATK Somatic SNV+Indel+CNV+SV(下)性能优化]]翻出来用一下。跑了一遍发现还是各种问题,于是想把pipeline改造成免部署、首次运行初始化环境的版本,以便需要时候能够直接运行起来,于是有了本文。
一句话描述就是:开箱即用的pipeline,能够根据样本version_reference自动选择参考基因组版本,根据project_bed文件选择项目bed,自动初始化环境、安装所需软件、下载ref文件和数据库的版本
为了让pipeline能够直接运行,无需部署,这里使用docker容器保证运行环境的一致性:见文章:基于docker的生信基础环境镜像构建,这里采用的方案是带ssh服务的docker+conda环境,整个pipeline在一个通用容器中运行。
本文代码较长,可能略复杂,想直接运行的可以下载 workflow文件,导入sliverworkspace图形化生信平台直接运行,获取并查看图形化分析报告
相关代码和workflow文件可以访问笔者github项目地址
导入workflow操作
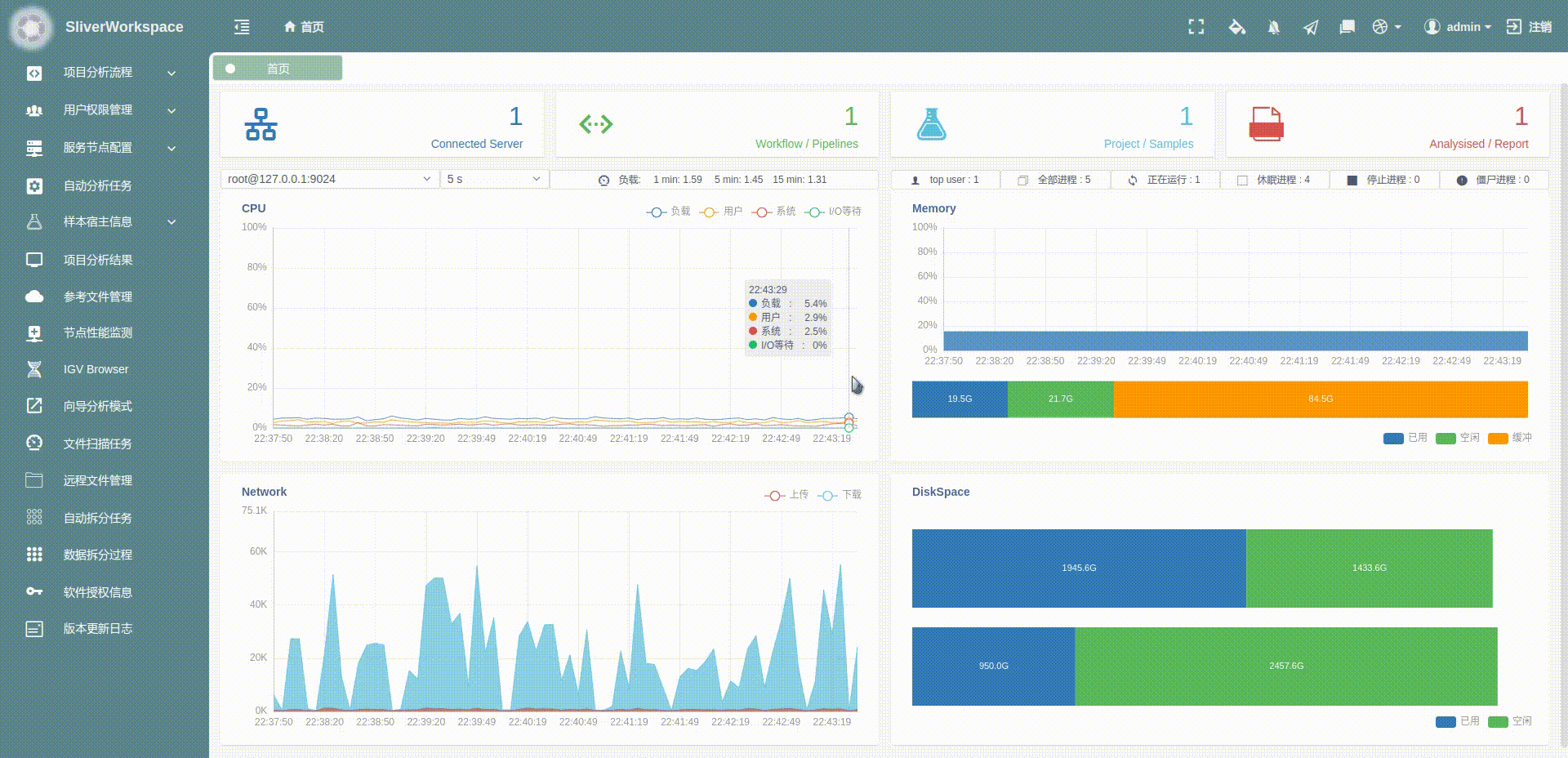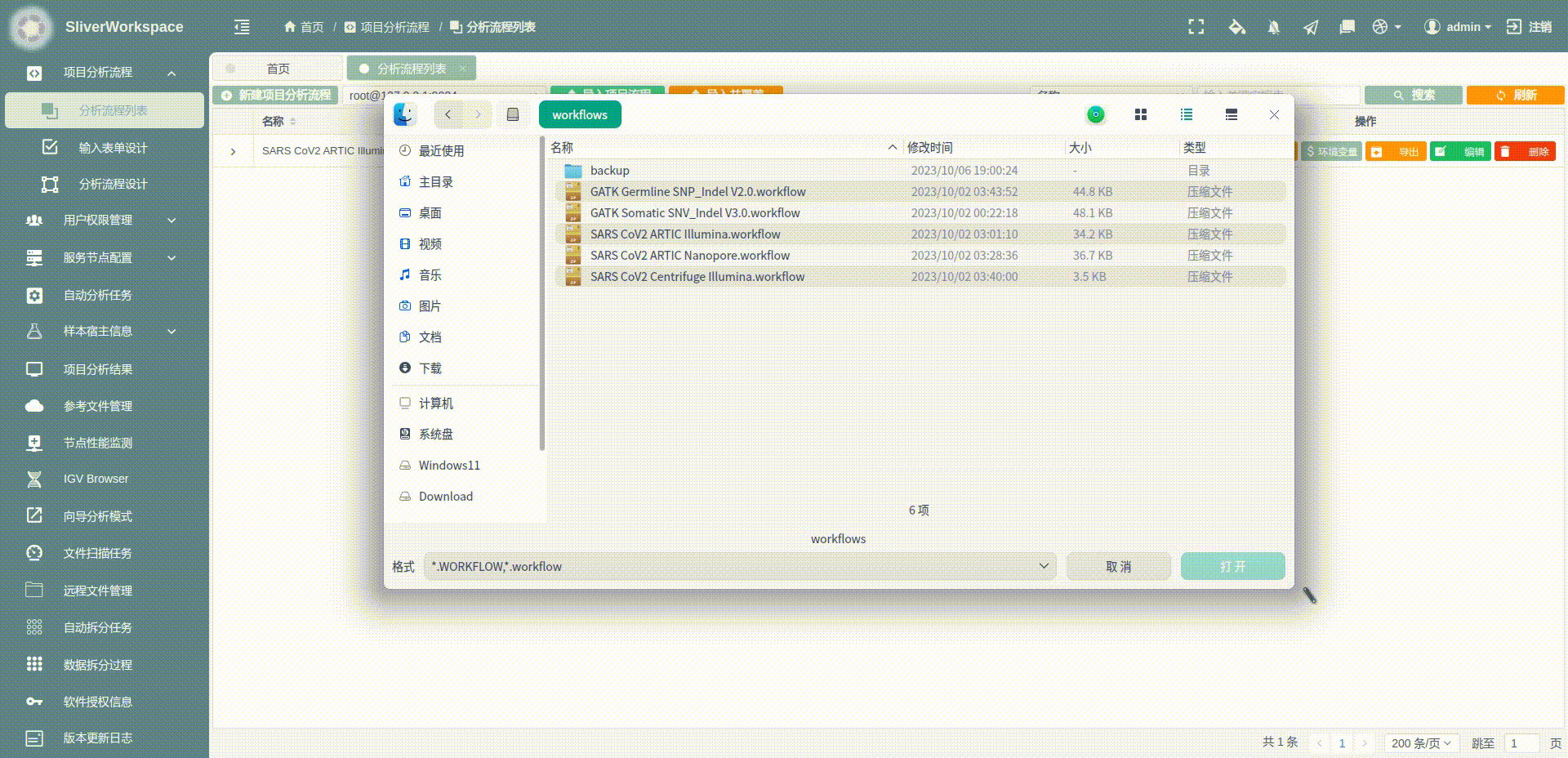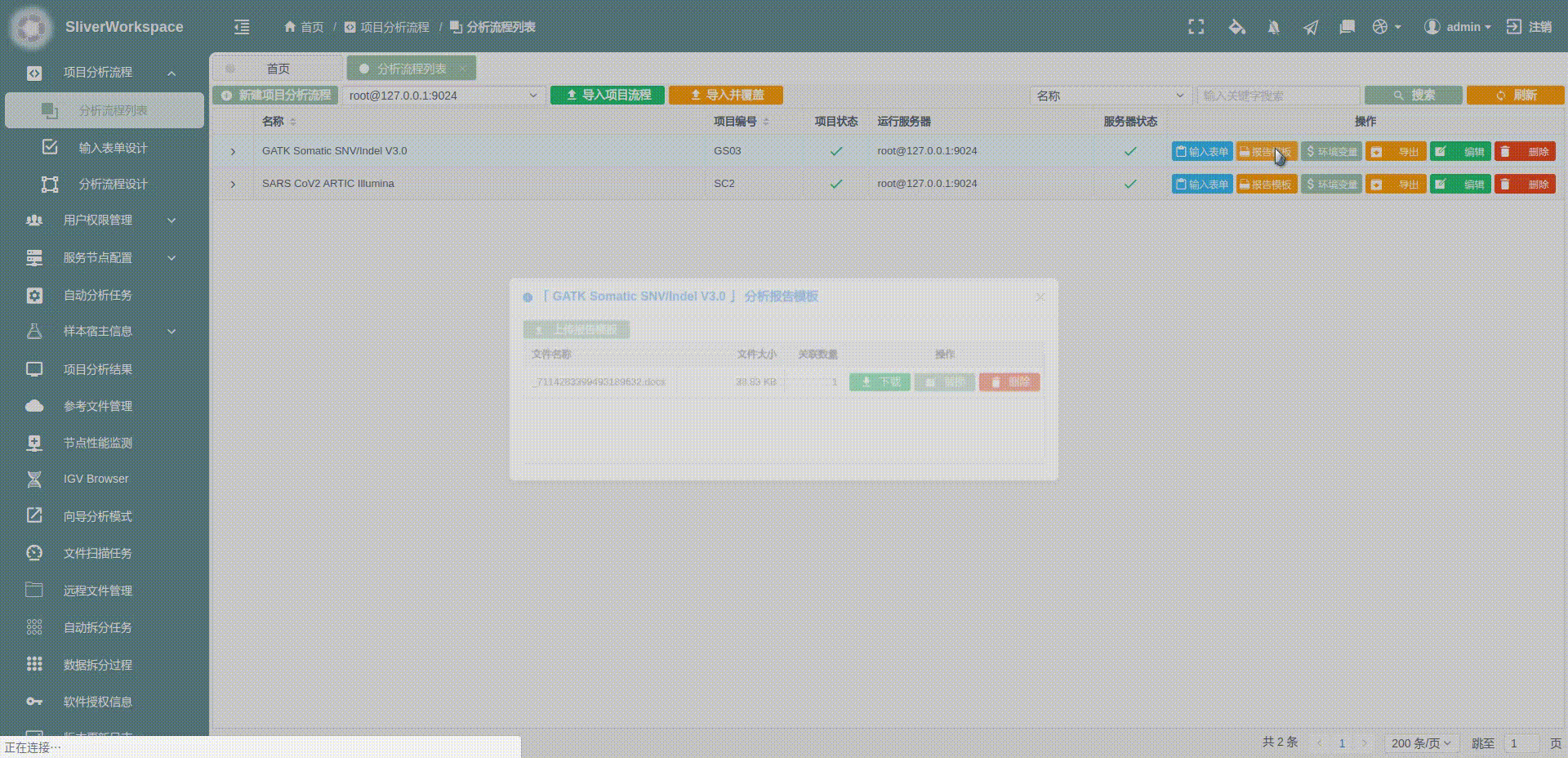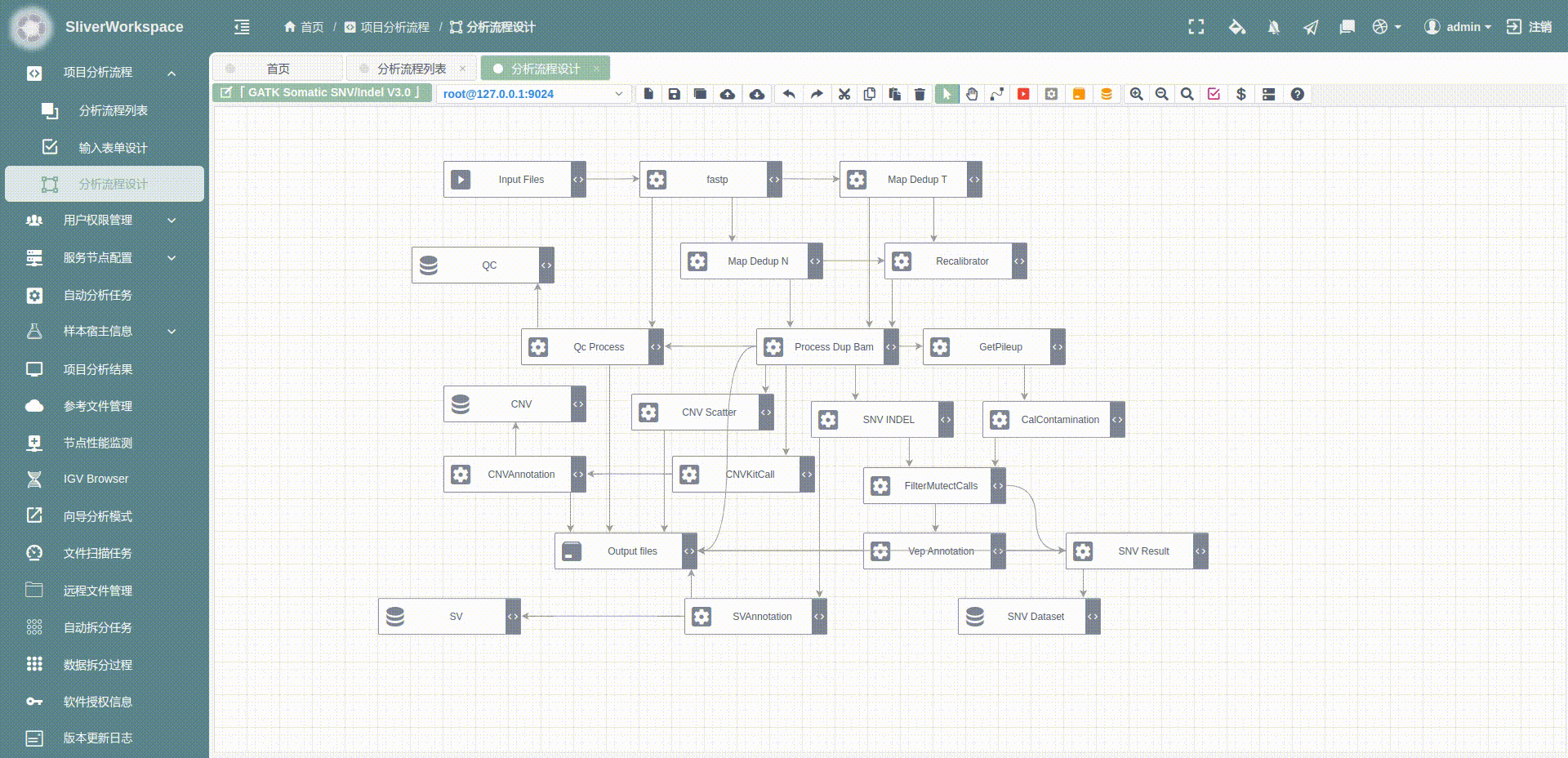
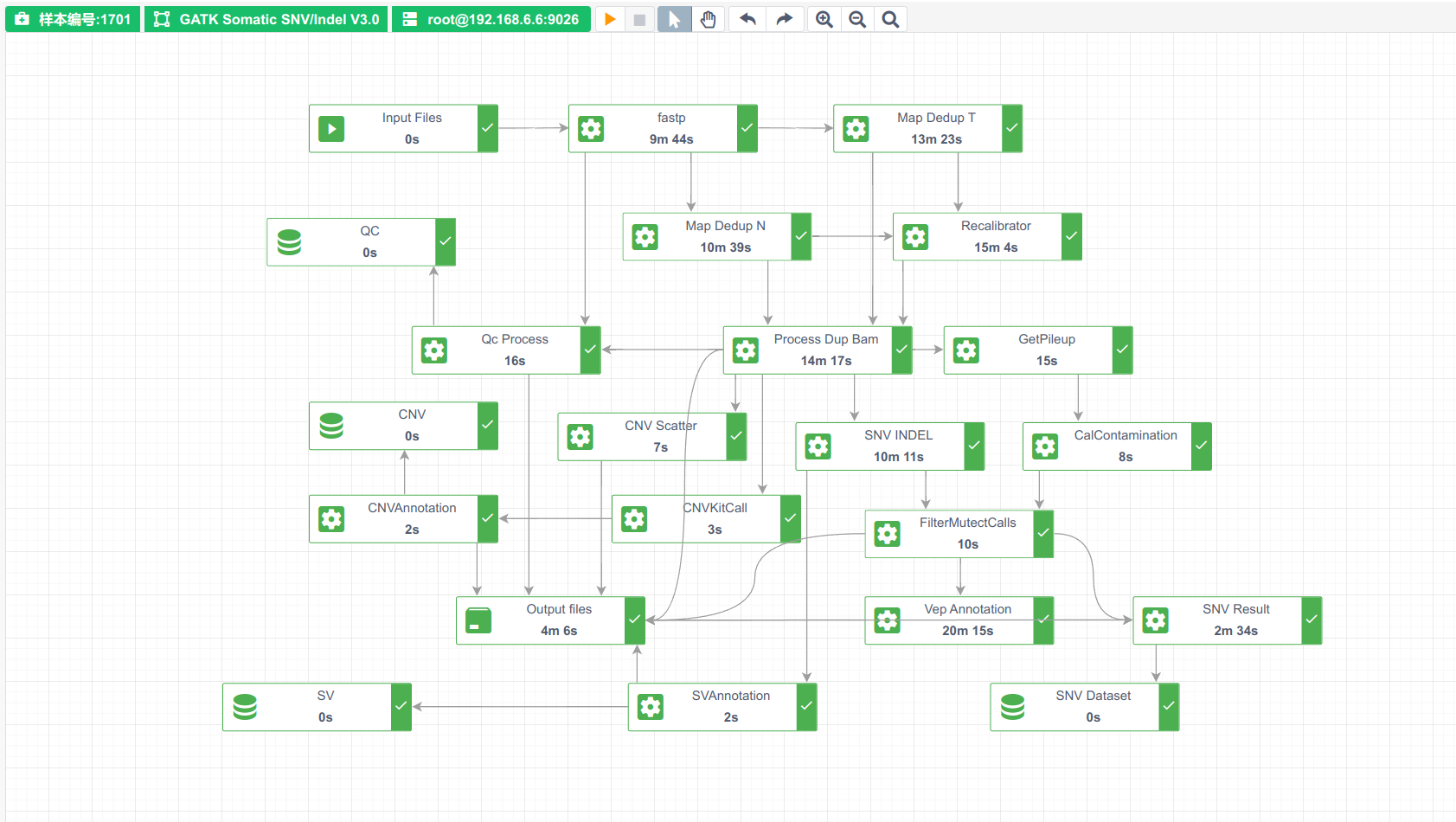
分析流程整体概览
docker 镜像拉取及部署配置
bash
# 拉取docker镜像
docker pull doujiangbaozi/sliverworkspace:1.10docker-compose.yml配置文件
yaml
version: "3"
services:
GATK:
image: doujiangbaozi/sliverworkspace:1.10
container_name: GATK
volumes:
- /home/sliver/Data/data:/opt/data:rw #挂载输入数据目录
- /home/sliver/Manufacture/gatk/envs:/root/mambaforge-pypy3/envs #挂载envs目录
- /home/sliver/Manufacture/sliver/ref:/opt/ref:rw #挂载reference目录
- /home/sliver/Manufacture/gatk/result:/opt/result:rw #挂载中间文件和结果目录
environment:
- TZ=Asia/Shanghai #设置时区
- PS=20191124 #设置ssh密码
- PT=9024 #设置ssh连接端口docker 镜像部署运行
bash
# 在docker-compose.yml文件同级目录下运行
docker-compose up -d
# 或者docker-compose -f docker-compose.yml所在目录
docker-compose -f somedir/docker-compose.yml up -d
# 可以通过ssh连接到docker 运行pipeline命令了,连接端口和密码见docker-compose.yml配置文件相关字段
ssh root@127.0.0.1 -p9024测试数据
测试数据来自2017年卫计委室间质评提供的bed文件(pipeline会自动下载)和测试数据,修改命名以匹配pipeline输入端,也可以替换为自己的数据文件,因为室间质评目前参考基因组还停留在hg19版本,所以本流程仍然使用hg19(GRCH37),如果要切换到hg38,可以将version_reference变量值设置为hg38,project_bed设置为Illumina_pt2_hg38.bed。pipeline会使用hg38(GRCH38)版本和对应的bed文件。
| 文件名(按照需要有调整) | 文件大小 | MD5 |
|---|---|---|
| B1701_R1.fq.gz | 4.85G | 07d3cdccee41dbb3adf5d2e04ab28e5b |
| B1701_R2.fq.gz | 4.77G | c2aa4a8ab784c77423e821b9f7fb00a7 |
| B1701NC_R1.fq.gz | 3.04G | 4fc21ad05f9ca8dc93d2749b8369891b |
| B1701NC_R2.fq.gz | 3.11G | bc64784f2591a27ceede1727136888b9 |
变量名称
bash
# 变量初始化赋值
sn=1701 #样本编号
pn=GS03 #pipeline 代号
data=/opt/data #数据输入目录
result=/opt/result #数据输出、中间文件目录
project_bed=Illumina_pt2.bed #参考基因组hg19下的bed文件
version_reference=hg19 #人参考基因组版本hg19或者hg38
version_fastqc=0.12.1 #fastqc 版本
version_multiqc=1.21 #multiqc 版本
version_cnvkit=0.9.10 #cnvkit 版本
version_manta=1.6.0 #manta 版本
version_gatk=4.3.0.0 #gatk 版本
version_sambamba=1.0.1 #sambamba 版本
version_samtools=1.17 #samtools 版本
version_bwa=0.7.17 #bwa 版本
version_fastp=0.23.2 #fastp 版本
version_vep=108.2 #vep软件版本
version_vep_cache=108 #vep-cache数据库版本
envs=/root/mambaforge-pypy3/envs #mamba envs 目录
threads=32 #最大线程数
memory=32G #内存占用
scatter=8 #Mutect2 分拆并行运行interval list 份数
event=2 #gatk FilterMutectCalls --max-events-in-region 数值
snv_tlod=16.00 #snv 过滤参数 tload 值
snv_vaf=0.01 #snv 过滤参数 丰度/突变频率
snv_depth=500 #snv 过滤参数 支持reads数/depth 测序深度
cnv_dep=1000 #cnv 过滤参数 支持reads数/depth 测序深度
cnv_min=-0.5 #cnv 过滤参数 log2最小值
cnv_max=0.5 #cnv 过滤参数 log2 最大值
sv_score=200 #sv 过滤参数 score
## 以上变量个可以具体根据需求调整表格:
| 变量名 | 变量值 | 备注 |
|---|---|---|
| sn | 1701 | 样本编号 |
| pn | GS03 | pipeline 代号 GATK Somatic 03版本 |
| project_bed | Illumina_pt2.bed | 参考基因组hg19下的bed文件 |
| version_reference | hg19 | 人参考基因组版本hg19 / hg38 |
| version_fastqc | 0.12.1 | fastqc 版本 |
| version_multiqc | 1.21 | multiqc 版本 |
| version_cnvkit | 0.9.10 | cnvkit 版本 |
| version_manta | 1.6.0 | manta版本 |
| version_gatk | 4.3.0.0 | gatk 版本 |
| version_sambamba | 1.0.1 | sambamba 版本 |
| version_samtools | 1.17 | samtools 版本 |
| version_bwa | 0.7.17 | bwa 版本 |
| version_fastp | 0.23.2 | fastp 版本 |
| version_vep | 108.2 | vep软件版本 |
| version_vep_cache | 108 | vep cache 版本 |
| envs | /root/mambaforge-pypy3/envs | mamba envs 目录 |
| threads | 32 | 最大线程数 |
| memory | 32G | 内存占用 |
| scatter | 8 | Mutect2 分拆并行运行interval list 份数 |
| event | 2 | gatk FilterMutectCalls --max-events-in-region 数值 |
| snv_tlod | 16.00 | snv 过滤参数 tload 值 |
| snv_vaf | 0.01 | snv 过滤参数 丰度/突变频率 |
| snv_depth | 500 | snv 过滤参数 支持reads数/depth 测序深度 |
| cnv_dep | 1000 | cnv 过滤参数 支持reads数/depth 测序深度 |
| cnv_min | -0.5 | cnv 过滤参数 log2最小值 |
| cnv_max | 0.5 | cnv 过滤参数 log2 最大值 |
| sv_score | 200 | sv 过滤参数 score |
Pipeline/workflow 具体步骤:
1. fastp 默认参数过滤,看下原始数据质量,clean data
bash
#conda检测环境是否存在,首次运行不存在创建该环境并安装软件
if [ ! -d "${envs}/${pn}.fastp" ]; then
echo "Creating the environment ${pn}.fastp"
mamba create -n ${pn}.fastp -y fastp=${version_fastp} fastqc=${version_fastqc} multiqc=${version_multiqc}
fi
mamba activate ${pn}.fastp
mkdir -p ${result}/${sn}/trimmed
mkdir -p ${result}/${sn}/qc
fastqc -t ${threads}\
${data}/GS03/${sn}_tumor_R1.fq.gz \
${data}/GS03/${sn}_tumor_R2.fq.gz \
-o ${result}/${sn}/qc &
fastqc -t ${threads}\
${data}/GS03/${sn}_normal_R1.fq.gz \
${data}/GS03/${sn}_normal_R2.fq.gz \
-o ${result}/${sn}/qc &
fastp -w 16 \
-i ${data}/GS03/${sn}_tumor_R1.fq.gz \
-I ${data}/GS03/${sn}_tumor_R2.fq.gz \
-o ${result}/${sn}/trimmed/${sn}_tumor_R1_trimmed.fq.gz \
-O ${result}/${sn}/trimmed/${sn}_tumor_R2_trimmed.fq.gz \
-h ${result}/${sn}/trimmed/${sn}_tumor_fastp.html \
-j ${result}/${sn}/trimmed/${sn}_tumor_fastp.json &
fastp -w 16 \
-i ${data}/GS03/${sn}_normal_R1.fq.gz \
-I ${data}/GS03/${sn}_normal_R2.fq.gz \
-o ${result}/${sn}/trimmed/${sn}_normal_R1_trimmed.fq.gz \
-O ${result}/${sn}/trimmed/${sn}_normal_R2_trimmed.fq.gz \
-h ${result}/${sn}/trimmed/${sn}_normal_fastp.html \
-j ${result}/${sn}/trimmed/${sn}_normal_fastp.json &
wait
fastqc -t ${threads}\
${result}/${sn}/trimmed/${sn}_tumor_R1_trimmed.fq.gz \
${result}/${sn}/trimmed/${sn}_tumor_R2_trimmed.fq.gz \
-o ${result}/${sn}/qc &
fastqc -t ${threads}\
${result}/${sn}/trimmed/${sn}_normal_R1_trimmed.fq.gz \
${result}/${sn}/trimmed/${sn}_normal_R2_trimmed.fq.gz \
-o ${result}/${sn}/qc &
wait
mamba deactivate2. normal文件fastq比对到参考基因组,sort 排序,mark duplicate 得到 marked.bam
bash
#conda检测环境是否存在,首次运行不存在创建该环境并安装软件
if [ ! -d "${envs}/${pn}.align" ]; then
mamba create -n ${pn}.align -y bwa=${version_bwa} samtools=${version_samtools}
fi
#从github下载sambamba static 比 mamba 安装的版本速度快1倍以上.这是个很诡异的地方
if [ ! -f "${envs}/${pn}.align/bin/sambamba" ]; then
aria2c https://github.com/biod/sambamba/releases/download/v${version_sambamba}/sambamba-${version_sambamba}-linux-amd64-static.gz -d ${envs}/${pn}.align/bin
gzip -cdf ${envs}/${pn}.align/bin/sambamba-${version_sambamba}-linux-amd64-static.gz > ${envs}/${pn}.align/bin/sambamba
chmod a+x ${envs}/${pn}.align/bin/sambamba
fi
mamba activate ${pn}.align
if [ "${version_reference}" == hg19 ]; then
echo "USE reference Version : ${version_reference}"
mkdir -p /opt/ref/hg19
#如果没有检测到参考基因组序列,则下载序列并使用bwa创建索引
if [ ! -f "/opt/ref/hg19/hg19.fasta" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg19/ucsc.hg19.fasta.gz -d /opt/ref/hg19 -o hg19.fasta.gz
cd /opt/ref/hg19 && gzip -d /opt/ref/hg19/hg19.fasta.gz
else
if [ -f "/opt/ref/hg19/ucsc.hg19.fasta.gz.aria2" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg19/ucsc.hg19.fasta.gz -c -d /opt/ref/hg19 -o hg19.fasta.gz
cd /opt/ref/hg19 && gzip -d /opt/ref/hg19/hg19.fasta.gz
fi
fi
if [ ! -f /opt/ref/hg19/hg19.fasta.amb ] ||
[ ! -f /opt/ref/hg19/hg19.fasta.ann ] ||
[ ! -f /opt/ref/hg19/hg19.fasta.bwt ] ||
[ ! -f /opt/ref/hg19/hg19.fasta.pac ] ||
[ ! -f /opt/ref/hg19/hg19.fasta.sa ]; then
bwa index /opt/ref/hg19/hg19.fasta
fi
#检测samtools索引是否存在,如不存在则使用samtools创建参考基因组索引
if [ ! -f "/opt/ref/hg19/hg19.fasta.fai" ]; then
samtools faidx /opt/ref/hg19/hg19.fasta
fi
elif [ "${version_reference}" == "hg38" ]; then
echo "USE reference Version : ${version_reference}"
mkdir -p /opt/ref/hg38
#如果没有检测到参考基因组序列,则下载序列并使用bwa创建索引
if [ ! -f "/opt/ref/hg38/hg38.fasta" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/Homo_sapiens_assembly38.fasta.gz -d /opt/ref/hg38 -o hg38.fasta.gz
cd /opt/ref/hg38 && gzip -d /opt/ref/hg38/hg38.fasta.gz
else
if [ -f "/opt/ref/hg38/hg38.fasta.gz.aria2" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/Homo_sapiens_assembly38.fasta.gz -d /opt/ref/hg38 -d /opt/ref/hg38 -o hg38.fasta.gz
cd /opt/ref/hg38 && gzip -d /opt/ref/hg38/hg38.fasta.gz
fi
fi
if [ ! -f /opt/ref/hg38/hg38.fasta.amb ] ||
[ ! -f /opt/ref/hg38/hg38.fasta.ann ] ||
[ ! -f /opt/ref/hg38/hg38.fasta.bwt ] ||
[ ! -f /opt/ref/hg38/hg38.fasta.pac ] ||
[ ! -f /opt/ref/hg38/hg38.fasta.sa ]; then
bwa index /opt/ref/hg38/hg38.fasta
fi
#检测samtools索引是否存在,如不存在则使用samtools创建参考基因组索引
if [ ! -f "/opt/ref/hg38/hg38.fasta.fai" ]; then
samtools faidx /opt/ref/hg38/hg38.fasta
fi
fi
mkdir -p ${result}/${sn}/aligned
#比对基因组管道输出成bam文件,管道输出排序
bwa mem \
-t ${threads} -M \
-R "@RG\\tID:${sn}_normal\\tLB:${sn}_normal\\tPL:Illumina\\tPU:Miseq\\tSM:${sn}_normal" \
/opt/ref/${version_reference}/${version_reference}.fasta ${result}/${sn}/trimmed/${sn}_normal_R1_trimmed.fq.gz ${result}/${sn}/trimmed/${sn}_normal_R2_trimmed.fq.gz \
| sambamba view -S -f bam -l 0 /dev/stdin \
| sambamba sort -t ${threads} -m 2G --tmpdir=${result}/${sn}/aligned -o ${result}/${sn}/aligned/${sn}_normal_sorted.bam /dev/stdin
#防止linux打开文件句柄数超过限制,报错退出
ulimit -n 10240
#使用sambamba对sorted bam文件标记重复
sambamba markdup \
--tmpdir ${result}/${sn}/aligned \
-t ${threads} ${result}/${sn}/aligned/${sn}_normal_sorted.bam ${result}/${sn}/aligned/${sn}_normal_marked.bam
#修改marked bam文件索引名,gatk和sambamba索引文件名需要保持一致
mv ${result}/${sn}/aligned/${sn}_normal_marked.bam.bai ${result}/${sn}/aligned/${sn}_normal_marked.bai
#删除sorted bam文件
rm -f ${result}/${sn}/aligned/${sn}_normal_sorted.bam*
mamba deactivate3. tumor文件fastq比对到参考基因组,排序,mark duplicate 得到 marked.bam,与第2步类似
bash
if [ ! -d "${envs}/${pn}.align" ]; then
mamba create -n ${pn}.align -y bwa=${version_bwa} samtools=${version_samtools}
fi
#从github下载sambamba static 比 mamba 安装的版本速度快1倍以上.
if [ ! -f "${envs}/${pn}.align/bin/sambamba" ]; then
aria2c https://github.com/biod/sambamba/releases/download/v${version_sambamba}/sambamba-${version_sambamba}-linux-amd64-static.gz -d ${envs}/${pn}.align/bin
gzip -cdf ${envs}/${pn}.align/bin/sambamba-${version_sambamba}-linux-amd64-static.gz > ${envs}/${pn}.align/bin/sambamba
chmod a+x ${envs}/${pn}.align/bin/sambamba
fi
mamba activate ${pn}.align
if [ "${version_reference}" == hg19 ]; then
echo "USE reference Version : ${version_reference}"
mkdir -p /opt/ref/hg19
#如果没有检测到参考基因组序列,则下载序列并使用bwa创建索引
if [ ! -f "/opt/ref/hg19/hg19.fasta" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg19/ucsc.hg19.fasta.gz -d /opt/ref/hg19 -o hg19.fasta.gz
cd /opt/ref/hg19 && gzip -d /opt/ref/hg19/hg19.fasta.gz
else
if [ -f "/opt/ref/hg19/ucsc.hg19.fasta.gz.aria2" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg19/ucsc.hg19.fasta.gz -c -d /opt/ref/hg19 -o hg19.fasta.gz
cd /opt/ref/hg19 && gzip -d /opt/ref/hg19/hg19.fasta.gz
fi
fi
if [ ! -f /opt/ref/hg19/hg19.fasta.amb ] ||
[ ! -f /opt/ref/hg19/hg19.fasta.ann ] ||
[ ! -f /opt/ref/hg19/hg19.fasta.bwt ] ||
[ ! -f /opt/ref/hg19/hg19.fasta.pac ] ||
[ ! -f /opt/ref/hg19/hg19.fasta.sa ]; then
bwa index /opt/ref/hg19/hg19.fasta
fi
#检测samtools索引是否存在,如不存在则使用samtools创建参考基因组索引
if [ ! -f "/opt/ref/hg19/hg19.fasta.fai" ]; then
samtools faidx /opt/ref/hg19/hg19.fasta
fi
elif [ "${version_reference}" == "hg38" ]; then
echo "USE reference Version : ${version_reference}"
mkdir -p /opt/ref/hg38
#如果没有检测到参考基因组序列,则下载序列并使用bwa创建索引
if [ ! -f "/opt/ref/hg38/hg38.fasta" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/Homo_sapiens_assembly38.fasta.gz -d /opt/ref/hg38 -o hg38.fasta.gz
cd /opt/ref/hg38 && gzip -d /opt/ref/hg38/hg38.fasta.gz
else
if [ -f "/opt/ref/hg38/hg38.fasta.gz.aria2" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/Homo_sapiens_assembly38.fasta.gz -d /opt/ref/hg38 -d /opt/ref/hg38 -o hg38.fasta.gz
cd /opt/ref/hg38 && gzip -d /opt/ref/hg38/hg38.fasta.gz
fi
fi
if [ ! -f /opt/ref/hg38/hg38.fasta.amb ] ||
[ ! -f /opt/ref/hg38/hg38.fasta.ann ] ||
[ ! -f /opt/ref/hg38/hg38.fasta.bwt ] ||
[ ! -f /opt/ref/hg38/hg38.fasta.pac ] ||
[ ! -f /opt/ref/hg38/hg38.fasta.sa ]; then
bwa index /opt/ref/hg38/hg38.fasta
fi
#检测samtools索引是否存在,如不存在则使用samtools创建参考基因组索引
if [ ! -f "/opt/ref/hg38/hg38.fasta.fai" ]; then
samtools faidx /opt/ref/hg38/hg38.fasta
fi
fi
mkdir -p ${result}/${sn}/aligned
bwa mem \
-t ${threads} -M \
-R "@RG\\tID:${sn}_tumor\\tLB:${sn}_tumor\\tPL:Illumina\\tPU:Miseq\\tSM:${sn}_tumor" \
/opt/ref/${version_reference}/${version_reference}.fasta ${result}/${sn}/trimmed/${sn}_tumor_R1_trimmed.fq.gz ${result}/${sn}/trimmed/${sn}_tumor_R2_trimmed.fq.gz \
| sambamba view -S -f bam -l 0 /dev/stdin \
| sambamba sort -t ${threads} -m 2G --tmpdir=${result}/${sn}/aligned -o ${result}/${sn}/aligned/${sn}_tumor_sorted.bam /dev/stdin
ulimit -n 10240
sambamba markdup \
--tmpdir ${result}/${sn}/aligned \
-t ${threads} ${result}/${sn}/aligned/${sn}_tumor_sorted.bam ${result}/${sn}/aligned/${sn}_tumor_marked.bam
mv ${result}/${sn}/aligned/${sn}_tumor_marked.bam.bai ${result}/${sn}/aligned/${sn}_tumor_marked.bai
rm -f ${result}/${sn}/aligned/${sn}_tumor_sorted.bam*
mamba deactivate
4. 对上述bam文件生成重新校准表,为后续BQSR使用;Generates recalibration table for Base Quality Score Recalibration (BQSR)
bash
#conda检测环境是否存在,首次运行不存在创建该环境并安装软件
if [ ! -f "${envs}/${pn}.gatk/bin/gatk" ]; then
mamba create -n ${pn}.gatk -y gatk4=${version_gatk} \
r-base=3.6.2 \
r-data.table=1.12.8 \
r-dplyr=0.8.5 \
r-getopt=1.20.3 \
r-ggplot2=3.3.0 \
r-gplots=3.0.3 \
r-gsalib=2.1 \
r-optparse=1.6.4 \
r-backports=1.1.10 \
tabix
fi
mamba activate ${pn}.gatk
if [ "${version_reference}" == hg19 ]; then
echo "USE reference Version : ${version_reference}"
#这里有个巨坑,broadinstitute ftp站点bundle目录提供的hg19版本参考文件,默认格式运行会报错,提示没有索引,使用gatk创建索引仍然报错,其实是gz格式需要使用bgzip重新压缩并且使用tabix创建索引才行
if [ ! -f "/opt/ref/hg19/dbsnp_138.hg19.vcf.gz" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg19/dbsnp_138.hg19.vcf.gz -d /opt/ref/hg19
else
if [ -f "/opt/ref/hg19/dbsnp_138.hg19.vcf.gz.aria2" ]; then
echo 'download continue...'
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg19/dbsnp_138.hg19.vcf.gz -c -d /opt/ref/hg19
fi
fi
if [ ! -f "/opt/ref/hg19/dbsnp_138.hg19.vcf.gz.tbi" ]; then
gzip -k -f -d /opt/ref/hg19/dbsnp_138.hg19.vcf.gz > /opt/ref/hg19/dbsnp_138.hg19.vcf
bgzip -f --threads ${threads} /opt/ref/hg19/dbsnp_138.hg19.vcf > /opt/ref/hg19/dbsnp_138.hg19.vcf.gz
tabix -f /opt/ref/hg19/dbsnp_138.hg19.vcf.gz
fi
if [ ! -f "/opt/ref/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf.gz" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf.gz -d /opt/ref/hg19
else
if [ -f "/opt/ref/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf.gz.aria2" ]; then
echo 'download continue...'
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf.gz -c -d /opt/ref/hg19
fi
fi
if [ ! -f "/opt/ref/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf.gz.tbi" ]; then
gzip -k -f -d /opt/ref/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf.gz > /opt/ref/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf
bgzip -f --threads ${threads} /opt/ref/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf > /opt/ref/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf.gz
tabix -f /opt/ref/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf.gz
fi
if [ ! -f "/opt/ref/hg19/1000G_phase1.snps.high_confidence.hg19.sites.vcf.gz" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg19/1000G_phase1.snps.high_confidence.hg19.sites.vcf.gz -d /opt/ref/hg19
else
if [ -f "/opt/ref/hg19/1000G_phase1.snps.high_confidence.hg19.sites.vcf.gz.aria2" ]; then
echo 'download continue...'
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg19/1000G_phase1.snps.high_confidence.hg19.sites.vcf.gz -c -d /opt/ref/hg19
fi
fi
if [ ! -f "/opt/ref/hg19/1000G_phase1.snps.high_confidence.hg19.sites.vcf.gz.tbi" ]; then
gzip -k -f -d /opt/ref/hg19/1000G_phase1.snps.high_confidence.hg19.sites.vcf.gz > /opt/ref/hg19/1000G_phase1.snps.high_confidence.hg19.sites.vcf
bgzip -f --threads ${threads} /opt/ref/hg19/1000G_phase1.snps.high_confidence.hg19.sites.vcf > /opt/ref/hg19/1000G_phase1.snps.high_confidence.hg19.sites.vcf.gz
tabix -f /opt/ref/hg19/1000G_phase1.snps.high_confidence.hg19.sites.vcf.gz
fi
if [ ! -f "/opt/ref/hg19/1000G_phase1.indels.hg19.sites.vcf.gz" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg19/1000G_phase1.indels.hg19.sites.vcf.gz -d /opt/ref/
else
if [ -f "/opt/ref/hg19/1000G_phase1.indels.hg19.sites.vcf.gz.aria2" ]; then
echo 'download continue...'
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg19/1000G_phase1.indels.hg19.sites.vcf.gz -c -d /opt/ref/hg19
fi
fi
if [ ! -f "/opt/ref/hg19/1000G_phase1.indels.hg19.sites.vcf.gz.tbi" ]; then
gzip -k -f -d /opt/ref/hg19/1000G_phase1.indels.hg19.sites.vcf.gz > /opt/ref/hg19/1000G_phase1.indels.hg19.sites.vcf
bgzip -f --threads ${threads} /opt/ref/hg19/1000G_phase1.indels.hg19.sites.vcf > /opt/ref/hg19/1000G_phase1.indels.hg19.sites.vcf.gz
tabix -f /opt/ref/hg19/1000G_phase1.indels.hg19.sites.vcf.gz
fi
#创建参考序列hg19的dict字典文件
if [ ! -f "/opt/ref/hg19/hg19.dict" ]; then
gatk CreateSequenceDictionary -R /opt/ref/hg19/hg19.fasta -O /opt/ref/hg19/hg19.dict
fi
if [ ! -f "/opt/ref/${version_reference}/${project_bed}" ]; then
#根据下载的Illumina_pt2.bed 文件创建interval list文件,坐标转换,起始坐标0修改为1
#sed 's/chr//; s/\t/ /g' /opt/ref/hg19/Illumina_pt2.bed > /opt/ref/hg19/Illumina_pt2.processed.bed
mkdir -p /opt/ref/${version_reference}
aria2c https://raw.githubusercontent.com/doujiangbaozi/sliverworkspace-util/main/somatic/projects/${project_bed} -d /opt/ref/hg19
if [ ! -f "/opt/ref/hg19/${project_bed}.interval_list" ]; then
gatk BedToIntervalList \
-I /opt/ref/hg19/${project_bed} \
-SD /opt/ref/hg19/hg19.dict \
-O /opt/ref/hg19/${project_bed}.interval_list
fi
fi
mkdir -p ${result}/${sn}/recal
gatk BaseRecalibrator \
--known-sites /opt/ref/hg19/dbsnp_138.hg19.vcf.gz \
--known-sites /opt/ref/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf.gz \
--known-sites /opt/ref/hg19/1000G_phase1.indels.hg19.sites.vcf.gz \
-L /opt/ref/hg19/${project_bed}.interval_list \
-R /opt/ref/hg19/hg19.fasta \
-I ${result}/${sn}/aligned/${sn}_tumor_marked.bam \
-O ${result}/${sn}/recal/${sn}_tumor_recal.table &
gatk BaseRecalibrator \
--known-sites /opt/ref/hg19/dbsnp_138.hg19.vcf.gz \
--known-sites /opt/ref/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf.gz \
--known-sites /opt/ref/hg19/1000G_phase1.indels.hg19.sites.vcf.gz \
-L /opt/ref/hg19/${project_bed}.interval_list \
-R /opt/ref/hg19/hg19.fasta \
-I ${result}/${sn}/aligned/${sn}_normal_marked.bam \
-O ${result}/${sn}/recal/${sn}_normal_recal.table &
wait
elif [ "${version_reference}" == "hg38" ]; then
echo "USE reference Version : ${version_reference}"
if [ ! -f "/opt/ref/hg38/dbsnp_146.hg38.vcf.gz" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/dbsnp_146.hg38.vcf.gz -d /opt/ref/hg38
else
if [ -f "/opt/ref/hg38/dbsnp_146.hg38.vcf.gz.aria2" ]; then
echo 'download continue...'
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/dbsnp_146.hg38.vcf.gz -c -d /opt/ref/hg38
fi
fi
if [ ! -f "/opt/ref/hg38/dbsnp_146.hg38.vcf.gz.tbi" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/dbsnp_146.hg38.vcf.gz.tbi -d /opt/ref/hg38
else
if [ -f "/opt/ref/hg38/dbsnp_146.hg38.vcf.gz.tbi.aria2" ]; then
echo 'download continue...'
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/dbsnp_146.hg38.vcf.gz.tbi -c -d /opt/ref/hg38
fi
fi
if [ ! -f "/opt/ref/hg38/Mills_and_1000G_gold_standard.indels.hg38.vcf.gz" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/Mills_and_1000G_gold_standard.indels.hg38.vcf.gz -d /opt/ref/hg38
else
if [ -f "/opt/ref/hg38/Mills_and_1000G_gold_standard.indels.hg38.vcf.gz.aria2" ]; then
echo 'download continue...'
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/Mills_and_1000G_gold_standard.indels.hg38.vcf.gz -c -d /opt/ref/hg38
fi
fi
if [ ! -f "/opt/ref/hg38/Mills_and_1000G_gold_standard.indels.hg38.vcf.gz.tbi" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/Mills_and_1000G_gold_standard.indels.hg38.vcf.gz.tbi -d /opt/ref/hg38
else
if [ -f "/opt/ref/hg38/Mills_and_1000G_gold_standard.indels.hg38.vcf.gz.tbi.aria2" ]; then
echo 'download continue...'
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/Mills_and_1000G_gold_standard.indels.hg38.vcf.gz.tbi -c -d /opt/ref/hg38
fi
fi
if [ ! -f "/opt/ref/hg38/1000G_phase1.snps.high_confidence.hg38.vcf.gz" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/1000G_phase1.snps.high_confidence.hg38.vcf.gz -d /opt/ref/hg38
else
if [ -f "/opt/ref/hg38/1000G_phase1.snps.high_confidence.hg38.vcf.gz.aria2" ]; then
echo 'download continue...'
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/1000G_phase1.snps.high_confidence.hg38.vcf.gz -c -d /opt/ref/hg38
fi
fi
if [ ! -f "/opt/ref/hg38/1000G_phase1.snps.high_confidence.hg38.vcf.gz.tbi" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/1000G_phase1.snps.high_confidence.hg38.vcf.gz.tbi -d /opt/ref/hg38
else
if [ -f "/opt/ref/hg38/1000G_phase1.snps.high_confidence.hg38.vcf.gz.tbi.aria2" ]; then
echo 'download continue...'
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/1000G_phase1.snps.high_confidence.hg38.vcf.gz.tbi -c -d /opt/ref/hg38
fi
fi
#创建参考序列hg38的dict字典文件
if [ ! -f "/opt/ref/hg38/hg38.dict" ]; then
gatk CreateSequenceDictionary -R /opt/ref/hg38/hg38.fasta -O /opt/ref/hg38/hg38.dict
fi
if [ ! -f "/opt/ref/${version_reference}/${project_bed}" ]; then
#根据下载的Illumina_pt2.bed 文件创建interval list文件,坐标转换,起始坐标0修改为1
#sed 's/chr//; s/\t/ /g' /opt/ref/hg38/Illumina_pt2.bed > /opt/ref/hg38/Illumina_pt2.processed.bed
mkdir -p /opt/ref/hg38
aria2c https://raw.githubusercontent.com/doujiangbaozi/sliverworkspace-util/main/somatic/projects/${project_bed} -d /opt/ref/hg38 -o ${project_bed}
if [ ! -f "/opt/ref/hg38/${project_bed}.interval_list" ]; then
gatk BedToIntervalList \
-I /opt/ref/hg38/${project_bed} \
-SD /opt/ref/hg38/hg38.dict \
-O /opt/ref/hg38/${project_bed}.interval_list
fi
fi
mkdir -p ${result}/${sn}/recal
gatk BaseRecalibrator \
--known-sites /opt/ref/hg38/dbsnp_146.hg38.vcf.gz \
--known-sites /opt/ref/hg38/Mills_and_1000G_gold_standard.indels.hg38.vcf.gz \
--known-sites /opt/ref/hg38/1000G_phase1.snps.high_confidence.hg38.vcf.gz \
-L /opt/ref/hg38/${project_bed}.interval_list \
-R /opt/ref/hg38/hg38.fasta \
-I ${result}/${sn}/aligned/${sn}_tumor_marked.bam \
-O ${result}/${sn}/recal/${sn}_tumor_recal.table &
gatk BaseRecalibrator \
--known-sites /opt/ref/hg38/dbsnp_146.hg38.vcf.gz \
--known-sites /opt/ref/hg38/Mills_and_1000G_gold_standard.indels.hg38.vcf.gz \
--known-sites /opt/ref/hg38/1000G_phase1.snps.high_confidence.hg38.vcf.gz \
-L /opt/ref/hg38/${project_bed}.interval_list \
-R /opt/ref/hg38/hg38.fasta \
-I ${result}/${sn}/aligned/${sn}_normal_marked.bam \
-O ${result}/${sn}/recal/${sn}_normal_recal.table &
wait
fi
mamba deactivate
5. 使用校准表对bam碱基质量校准,因为这一步gatk效率感人,所以同时计算insertsize,拆分interval list(后续mutect2并行运行需要),运行cnvkit batch,运行samtools depth计算测序深度,samtools flagstat 统计mapping比例及质量
bash
mkdir -p ${result}/${sn}/bqsr
mkdir -p ${result}/${sn}/stat
mkdir -p ${result}/${sn}/cnv
mkdir -p ${result}/${sn}/interval
mamba activate ${pn}.gatk
gatk ApplyBQSR \
--bqsr-recal-file ${result}/${sn}/recal/${sn}_tumor_recal.table \
-L /opt/ref/${version_reference}/${project_bed}.interval_list \
-R /opt/ref/${version_reference}/${version_reference}.fasta \
-I ${result}/${sn}/aligned/${sn}_tumor_marked.bam \
-O ${result}/${sn}/bqsr/${sn}_tumor_bqsr.bam &
gatk ApplyBQSR \
--bqsr-recal-file ${result}/${sn}/recal/${sn}_normal_recal.table \
-L /opt/ref/${version_reference}/${project_bed}.interval_list \
-R /opt/ref/${version_reference}/${version_reference}.fasta \
-I ${result}/${sn}/aligned/${sn}_normal_marked.bam \
-O ${result}/${sn}/bqsr/${sn}_normal_bqsr.bam &
gatk CollectInsertSizeMetrics \
-I ${result}/${sn}/aligned/${sn}_tumor_marked.bam \
-O ${result}/${sn}/stat/${sn}_tumor_insertsize_metrics.txt \
-H ${result}/${sn}/stat/${sn}_tumor_insertsize_histogram.pdf &
gatk CollectInsertSizeMetrics \
-I ${result}/${sn}/aligned/${sn}_normal_marked.bam \
-O ${result}/${sn}/stat/${sn}_normal_insertsize_metrics.txt \
-H ${result}/${sn}/stat/${sn}_normal_insertsize_histogram.pdf &
rm -f ${result}/${sn}/interval/*.interval_list
gatk SplitIntervals \
-L /opt/ref/${version_reference}/${project_bed}.interval_list \
-R /opt/ref/${version_reference}/${version_reference}.fasta \
-O ${result}/${sn}/interval \
--scatter-count ${scatter} &
mamba deactivate
if [ ! -d "${envs}/${pn}.cnvkit" ]; then
mamba create -n ${pn}.cnvkit -y cnvkit=${version_cnvkit}
fi
if [ ! -f "/opt/ref/${version_reference}/refFlat.txt" ]; then
aria2c -x 16 -s 16 http://hgdownload.soe.ucsc.edu/goldenPath/${version_reference}/database/refFlat.txt.gz -d /opt/ref/${version_reference}
cd /opt/ref/${version_reference} && gzip -d refFlat.txt.gz
fi
mamba activate ${pn}.cnvkit
rm -f ${result}/${sn}/cnv/${sn}_reference.cnn
cnvkit.py batch \
${result}/${sn}/aligned/${sn}_tumor_marked.bam \
--normal ${result}/${sn}/aligned/${sn}_normal_marked.bam \
--method hybrid \
--fasta /opt/ref/${version_reference}/${version_reference}.fasta \
--targets /opt/ref/${version_reference}/${project_bed} \
--annotate /opt/ref/${version_reference}/refFlat.txt \
--output-reference ${result}/${sn}/cnv/${sn}_reference.cnn \
--output-dir ${result}/${sn}/cnv/ \
--diagram \
-p ${threads} &
mamba deactivate
mamba activate ${pn}.align
samtools depth -a -b /opt/ref/${version_reference}/Illumina_pt2.bed --threads ${threads} \
${result}/${sn}/aligned/${sn}_tumor_marked.bam > \
${result}/${sn}/stat/${sn}_tumor_marked.depth &
samtools depth -a -b /opt/ref/${version_reference}/Illumina_pt2.bed --threads ${threads} \
${result}/${sn}/aligned/${sn}_normal_marked.bam > \
${result}/${sn}/stat/${sn}_normal_marked.depth &
samtools flagstat --threads ${threads} \
${result}/${sn}/aligned/${sn}_tumor_marked.bam > \
${result}/${sn}/stat/${sn}_tumor_marked.flagstat &
samtools flagstat --threads ${threads} \
${result}/${sn}/aligned/${sn}_normal_marked.bam > \
${result}/${sn}/stat/${sn}_normal_marked.flagstat &
mamba deactivate
wait
6. 计算堆叠数据( pileup metrics )以便后续评估污染,也可以根据拆分的interval list并行处理,处理之后合并。
bash
#官方巨坑,默认提供的small_exac_common_3_b37.vcf.gz默认染色体坐标不是以chr开头而是数字
mamba activate ${pn}.gatk
if [ "${version_reference}" == hg19 ]; then
echo "USE reference Version : ${version_reference}"
#这里有个巨坑,从broadinstitute ftp 站点bundle Mutect2目录下载的参考文件,与同样下载的参考序列基因组坐标系不一致,参考基因组参考序列是chr1这种格式,这个af-only-gnomad是1,2,3这种格式,需要编写脚本处理
if [ ! -f "/opt/ref/hg19/small_exac_common_3_b37.processed.vcf.gz" ]; then
aria2c ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/Mutect2/GetPileupSummaries/small_exac_common_3_b37.vcf.gz -d /opt/ref/hg19
else
if [ -f "/opt/ref/hg19/small_exac_common_3_b37.vcf.gz.aria2" ]; then
aria2c ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/Mutect2/GetPileupSummaries/small_exac_common_3_b37.vcf.gz -c -d /opt/ref/hg19
fi
fi
if [ ! -f "/opt/ref/${version_reference}/small_exac_common_3_b37.processed.vcf.gz" ]; then
if [ ! -f "${envs}/VcfProcessUtil.py" ]; then
aria2c https://raw.fgit.cf/doujiangbaozi/sliverworkspace-util/main/somatic/VcfProcessUtil.py -d ${envs}/
#aria2c https://raw.githubusercontent.com/doujiangbaozi/sliverworkspace-util/main/somatic/VcfProcessUtil.py -d ${envs}/
chmod a+x ${envs}/VcfProcessUtil.py
fi
mamba activate ${pn}.cnvkit
${envs}/VcfProcessUtil.py \
-f /opt/ref/${version_reference}/small_exac_common_3_b37.vcf.gz \
-o /opt/ref/${version_reference}/small_exac_common_3_b37.processed.vcf
mamba deactivate
mamba activate ${pn}.gatk
cd /opt/ref/${version_reference}
bgzip -f --threads ${threads} small_exac_common_3_b37.processed.vcf
tabix -f small_exac_common_3_b37.processed.vcf.gz
mamba deactivate
fi
mamba activate ${pn}.gatk
for i in `ls ${result}/${sn}/interval/*.interval_list`;
do
echo $i
gatk GetPileupSummaries \
-R /opt/ref/${version_reference}/${version_reference}.fasta \
-I ${result}/${sn}/bqsr/${sn}_tumor_bqsr.bam \
-O ${i%.*}-tumor-pileups.table \
-V /opt/ref/${version_reference}/small_exac_common_3_b37.processed.vcf.gz \
-L $i \
--interval-set-rule INTERSECTION &
gatk GetPileupSummaries \
-R /opt/ref/${version_reference}/${version_reference}.fasta \
-I ${result}/${sn}/bqsr/${sn}_normal_bqsr.bam \
-O ${i%.*}-normal-pileups.table \
-V /opt/ref/${version_reference}/small_exac_common_3_b37.processed.vcf.gz \
-L $i \
--interval-set-rule INTERSECTION &
done
wait
mamba deactivate
elif [ "${version_reference}" == "hg38" ]; then
echo "USE reference Version : ${version_reference}"
if [ ! -f "/opt/ref/${version_reference}/small_exac_common_3.hg38.vcf.gz" ]; then
aria2c ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/Mutect2/GetPileupSummaries/small_exac_common_3.hg38.vcf.gz -d /opt/ref/${version_reference}
else
if [ -f "/opt/ref/hg19/small_exac_common_3_b37.vcf.gz.aria2" ]; then
aria2c ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/Mutect2/GetPileupSummaries/small_exac_common_3.hg38.vcf.gz -c -d /opt/ref/${version_reference}
fi
fi
if [ ! -f "/opt/ref/${version_reference}/small_exac_common_3.hg38.vcf.gz.tbi" ]; then
aria2c ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/Mutect2/GetPileupSummaries/small_exac_common_3.hg38.vcf.gz.tbi -d /opt/ref/${version_reference}
else
if [ -f "/opt/ref/hg19/small_exac_common_3_b37.vcf.gz.tbi.aria2" ]; then
aria2c ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/Mutect2/GetPileupSummaries/small_exac_common_3.hg38.vcf.gz.tbi -c -d /opt/ref/${version_reference}
fi
fi
mamba activate ${pn}.gatk
for i in `ls ${result}/${sn}/interval/*.interval_list`;
do
echo $i
gatk GetPileupSummaries \
-R /opt/ref/${version_reference}/${version_reference}.fasta \
-I ${result}/${sn}/bqsr/${sn}_tumor_bqsr.bam \
-O ${i%.*}-tumor-pileups.table \
-V /opt/ref/${version_reference}/small_exac_common_3.hg38.vcf.gz \
-L $i \
--interval-set-rule INTERSECTION &
gatk GetPileupSummaries \
-R /opt/ref/${version_reference}/${version_reference}.fasta \
-I ${result}/${sn}/bqsr/${sn}_normal_bqsr.bam \
-O ${i%.*}-normal-pileups.table \
-V /opt/ref/${version_reference}/small_exac_common_3.hg38.vcf.gz \
-L $i \
--interval-set-rule INTERSECTION &
done
wait
mamba deactivate
fi
mamba activate ${pn}.gatk
tables=
for i in `ls ${result}/${sn}/interval/*-tumor-pileups.table`;
do
tables="$tables -I $i"
done
gatk GatherPileupSummaries \
--sequence-dictionary /opt/ref/${version_reference}/${version_reference}.dict \
$tables \
-O ${result}/${sn}/stat/${sn}_tumor_pileups.table
nctables=
for i in `ls ${result}/${sn}/interval/*-normal-pileups.table`;
do
nctables="$nctables -I $i"
done
gatk GatherPileupSummaries \
--sequence-dictionary /opt/ref/${version_reference}/${version_reference}.dict \
$nctables \
-O ${result}/${sn}/stat/${sn}_normal_pileups.table
mamba deactivate
7. 使用GetPileupSummaries计算结果评估跨样本污染,结果用于后面 FilterMutectCall 过滤Mutect2输出结果
bash
mamba activate ${pn}.gatk
gatk CalculateContamination \
-matched ${result}/${sn}/stat/${sn}_normal_pileups.table \
-I ${result}/${sn}/stat/${sn}_tumor_pileups.table \
-O ${result}/${sn}/stat/${sn}_contamination.table
mamba deactivate
8. Mutect2 call 突变,使用拆分的interval list,结束后将结果合并;同时并行运行manta call sv突变
bash
mkdir -p ${result}/${sn}/sv
mkdir -p ${result}/${sn}/snv
mamba activate ${pn}.gatk
if [ "${version_reference}" == hg19 ]; then
echo "USE reference Version : ${version_reference}"
#这里有个巨坑,从broadinstitute ftp 站点bundle Mutect2目录下载的参考文件,与同样下载的参考序列基因组坐标系不一致,参考基因组参考序列是chr1这种格式,这个af-only-gnomad是1,2,3这种格式,需要编写脚本处理;hg38貌似没有这个问题,hg19的数据都不维护了么?
if [ ! -f "/opt/ref/hg19/af-only-gnomad.raw.sites.b37.vcf.gz" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/Mutect2/af-only-gnomad.raw.sites.b37.vcf.gz -d /opt/ref/hg19
else
if [ -f "/opt/ref/hg19/af-only-gnomad.raw.sites.b37.vcf.gz.aria2" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/Mutect2/af-only-gnomad.raw.sites.b37.vcf.gz -c -d /opt/ref/hg19
fi
if [ ! -f "${envs}/VcfProcessUtil.py" ]; then
aria2c https://raw.fgit.cf/doujiangbaozi/sliverworkspace-util/main/somatic/VcfProcessUtil.py -d ${envs}/
#aria2c https://raw.githubusercontent.com/doujiangbaozi/sliverworkspace-util/main/somatic/VcfProcessUtil.py -d ${envs}/
chmod a+x ${envs}/VcfProcessUtil.py
fi
mamba activate ${pn}.cnvkit
${envs}/VcfProcessUtil.py \
-f /opt/ref/hg19/af-only-gnomad.raw.sites.b37.vcf.gz \
-o /opt/ref/hg19/af-only-gnomad.raw.sites.b37.processed.vcf
mamba deactivate
mamba activate ${pn}.gatk
cd /opt/ref/hg19
bgzip -f --threads ${threads} af-only-gnomad.raw.sites.b37.processed.vcf
tabix -f af-only-gnomad.raw.sites.b37.processed.vcf.gz
mamba deactivate
fi
elif [ "${version_reference}" == "hg38" ]; then
echo "USE reference Version : ${version_reference}"
if [ ! -f "/opt/ref/${version_reference}/af-only-gnomad.hg38.vcf.gz" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/Mutect2/af-only-gnomad.hg38.vcf.gz -d /opt/ref/${version_reference}
else
if [ -f "/opt/ref/${version_reference}/af-only-gnomad.hg38.vcf.gz.aria2" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/Mutect2/af-only-gnomad.hg38.vcf.gz -c -d /opt/ref/${version_reference}
fi
fi
if [ ! -f "/opt/ref/${version_reference}/af-only-gnomad.hg38.vcf.gz.tbi" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/Mutect2/af-only-gnomad.hg38.vcf.gz.tbi -d /opt/ref/${version_reference}
else
if [ -f "/opt/ref/${version_reference}/af-only-gnomad.hg38.vcf.gz.tbi.aria2" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/Mutect2/af-only-gnomad.hg38.vcf.gz.tbi -c -d /opt/ref/${version_reference}
fi
fi
fi
mamba activate ${pn}.gatk
if [ ! -f "/opt/ref/${version_reference}/${project_bed}.gz" ]; then
bgzip -f -c /opt/ref/${version_reference}/${project_bed} > /opt/ref/hg19/${project_bed}.gz
tabix -f -p bed /opt/ref/${version_reference}/${project_bed}.gz
else
if [ ! -f "/opt/ref/${version_reference}/${project_bed}.gz.tbi" ]; then
tabix -f -p bed /opt/ref/${version_reference}/${project_bed}.gz
fi
fi
mamba deactivate
if [ ! -d "${envs}/${pn}.manta" ]; then
mamba create -n ${pn}.manta -y manta=1.6.0
fi
mamba activate ${pn}.manta
rm -f ${result}/${sn}/sv/runWorkflow.py*
configManta.py \
--normalBam ${result}/${sn}/bqsr/${sn}_normal_bqsr.bam \
--tumorBam ${result}/${sn}/bqsr/${sn}_tumor_bqsr.bam \
--referenceFasta /opt/ref/${version_reference}/${version_reference}.fasta \
--exome \
--callRegions /opt/ref/${version_reference}/${project_bed}.gz \
--runDir ${result}/${sn}/sv
rm -rf ${result}/${sn}/sv/workspace
python ${result}/${sn}/sv/runWorkflow.py -m local -j ${threads} &
mamba deactivate
mamba activate ${pn}.gatk
if [ "${version_reference}" == hg19 ]; then
echo "USE reference Version : ${version_reference}"
rm -f ${result}/${sn}/snv/vcf-file.list
touch ${result}/${sn}/snv/vcf-file.list
for i in `ls ${result}/${sn}/interval/*.interval_list`;
do
rm -f ${i%.*}_bqsr.vcf.gz
gatk Mutect2 \
-R /opt/ref/${version_reference}/${version_reference}.fasta \
-I ${result}/${sn}/bqsr/${sn}_tumor_bqsr.bam -tumor ${sn}_tumor \
-I ${result}/${sn}/bqsr/${sn}_normal_bqsr.bam -normal ${sn}_normal \
-L $i \
-O ${i%.*}_bqsr.vcf.gz \
--max-mnp-distance 10 \
--germline-resource /opt/ref/${version_reference}/af-only-gnomad.raw.sites.b37.processed.vcf.gz \
--native-pair-hmm-threads ${threads} &
echo ${i%.*}_bqsr.vcf.gz >> ${result}/${sn}/snv/vcf-file.list
done
elif [ "${version_reference}" == "hg38" ]; then
echo "USE reference Version : ${version_reference}"
rm -f ${result}/${sn}/snv/vcf-file.list
touch ${result}/${sn}/snv/vcf-file.list
for i in `ls ${result}/${sn}/interval/*.interval_list`;
do
rm -f ${i%.*}_bqsr.vcf.gz
gatk Mutect2 \
-R /opt/ref/${version_reference}/${version_reference}.fasta \
-I ${result}/${sn}/bqsr/${sn}_tumor_bqsr.bam -tumor ${sn}_tumor \
-I ${result}/${sn}/bqsr/${sn}_normal_bqsr.bam -normal ${sn}_normal \
-L $i \
-O ${i%.*}_bqsr.vcf.gz \
--max-mnp-distance 10 \
--germline-resource /opt/ref/${version_reference}/af-only-gnomad.hg38.vcf.gz \
--native-pair-hmm-threads ${threads} &
echo ${i%.*}_bqsr.vcf.gz >> ${result}/${sn}/snv/vcf-file.list
done
fi
wait
rm -f ${result}/${sn}/snv/${sn}_bqsr.vcf.gz.stats
stats=
for z in `ls ${result}/${sn}/interval/*_bqsr.vcf.gz.stats`;
do
stats="$stats -stats $z"
done
gatk MergeMutectStats $stats \
-O ${result}/${sn}/snv/${sn}_bqsr.vcf.gz.stats
gatk MergeVcfs \
-I ${result}/${sn}/snv/vcf-file.list \
-O ${result}/${sn}/snv/${sn}_bqsr.vcf.gz
mamba deactivate
9. FilterMutectCalls 对Mutect结果突变过滤
bash
mamba activate ${pn}.gatk
gatk FilterMutectCalls \
--max-events-in-region ${event} \
--contamination-table ${result}/${sn}/stat/${sn}_contamination.table \
-R /opt/ref/${version_reference}/${version_reference}.fasta \
-V ${result}/${sn}/snv/${sn}_bqsr.vcf.gz \
-O ${result}/${sn}/snv/${sn}_filtered.vcf.gz
mamba deactivate
10. 使用Vep注释过滤结果
bash
#conda检测环境是否存在,首次运行不存在创建该环境并安装软件
if [ ! -d "${envs}/${pn}.vep" ]; then
echo "Creating the environment ${pn}.vep"
mamba create -n ${pn}.vep -y ensembl-vep=${version_vep}
fi
mkdir -p /opt/result/${sn}/vcf
if [ "${version_reference}" == hg19 ]; then
echo "USE reference Version : ${version_reference}"
#检测vep注释数据库是否存在如果不存在则先下载
if [ ! -d "/opt/ref/vep-cache/homo_sapiens/${version_vep_cache}_GRCh37" ]; then
aria2c -x 16 -s 48 https://ftp.ensembl.org/pub/release-${version_vep_cache}/variation/indexed_vep_cache/homo_sapiens_vep_${version_vep_cache}_GRCh37.tar.gz -d /opt/ref/
tar -zxvf /opt/ref/homo_sapiens_vep_${version_vep_cache}_GRCh37.tar.gz -C /opt/ref/vep-cache/
elif [ -f "/opt/ref/homo_sapiens_vep_${version_vep_cache}_GRCh37.tar.gz.aria2" ]; then
aria2c -x 16 -s 48 https://ftp.ensembl.org/pub/release-${version_vep_cache}/variation/indexed_vep_cache/homo_sapiens_vep_${version_vep_cache}_GRCh37.tar.gz -c -d /opt/ref/
tar -zxvf /opt/ref/homo_sapiens_vep_${version_vep_cache}_GRCh37.tar.gz -C /opt/ref/vep-cache/
fi
if [ ! -d "/opt/ref/vep-cache/homo_sapiens_refseq/${version_vep_cache}_GRCh37" ]; then
aria2c -x 16 -s 48 http://ftp.ensembl.org/pub/release-${version_vep_cache}/variation/vep/homo_sapiens_refseq_vep_${version_vep_cache}_GRCh37.tar.gz -d /opt/ref/
tar -zxvf /opt/ref/homo_sapiens_refseq_vep_${version_vep_cache}_GRCh37.tar.gz -C /opt/ref/vep-cache/
elif [ -f "/opt/ref/homo_sapiens_refseq_vep_${version_vep_cache}_GRCh37.tar.gz.aria2" ]; then
aria2c -x 16 -s 48 http://ftp.ensembl.org/pub/release-${version_vep_cache}/variation/vep/homo_sapiens_refseq_vep_${version_vep_cache}_GRCh37.tar.gz -c -d /opt/ref/
tar -zxvf /opt/ref/homo_sapiens_refseq_vep_${version_vep_cache}_GRCh37.tar.gz -C /opt/ref/vep-cache/
fi
mamba activate ${pn}.vep
mkdir -p ${result}/${sn}/annotation
vep \
-i ${result}/${sn}/snv/${sn}_filtered.vcf.gz \
-o ${result}/${sn}/annotation/${sn}_filtered_vep.tsv \
--offline \
--cache \
--cache_version ${version_vep_cache} \
--everything \
--dir_cache /opt/ref/vep-cache/ \
--dir_plugins /opt/ref/vep-cache/Plugins \
--species homo_sapiens \
--assembly GRCh37 \
--fasta /opt/ref/${version_reference}/${version_reference}.fasta \
--refseq \
--force_overwrite \
--format vcf \
--tab \
--shift_3prime 1 \
--offline
mamba deactivate
elif [ "${version_reference}" == "hg38" ]; then
echo "USE reference Version : ${version_reference}"
#检测vep注释数据库是否存在如果不存在则先下载
if [ ! -d "/opt/ref/vep-cache/homo_sapiens/${version_vep_cache}_GRCh38" ]; then
aria2c -x 16 -s 48 https://ftp.ensembl.org/pub/release-${version_vep_cache}/variation/indexed_vep_cache/homo_sapiens_vep_${version_vep_cache}_GRCh38.tar.gz -d /opt/ref/
tar -zxvf /opt/ref/homo_sapiens_vep_${version_vep_cache}_GRCh38.tar.gz -C /opt/ref/vep-cache/
elif [ -f "/opt/ref/homo_sapiens_vep_${version_vep_cache}_GRCh38.tar.gz.aria2" ]; then
aria2c -x 16 -s 48 https://ftp.ensembl.org/pub/release-${version_vep_cache}/variation/indexed_vep_cache/homo_sapiens_vep_${version_vep_cache}_GRCh38.tar.gz -c -d /opt/ref/
tar -zxvf /opt/ref/homo_sapiens_vep_${version_vep_cache}_GRCh38.tar.gz -C /opt/ref/vep-cache/
fi
if [ ! -d "/opt/ref/vep-cache/homo_sapiens_refseq/${version_vep_cache}_GRCh38" ]; then
aria2c -x 16 -s 48 http://ftp.ensembl.org/pub/release-${version_vep_cache}/variation/vep/homo_sapiens_refseq_vep_${version_vep_cache}_GRCh38.tar.gz -d /opt/ref/
tar -zxvf /opt/ref/homo_sapiens_refseq_vep_${version_vep_cache}_GRCh38.tar.gz -C /opt/ref/vep-cache/
elif [ -f "/opt/ref/homo_sapiens_refseq_vep_${version_vep_cache}_GRCh38.tar.gz.aria2" ]; then
aria2c -x 16 -s 48 http://ftp.ensembl.org/pub/release-${version_vep_cache}/variation/vep/homo_sapiens_refseq_vep_${version_vep_cache}_GRCh38.tar.gz -c -d /opt/ref/
tar -zxvf /opt/ref/homo_sapiens_refseq_vep_${version_vep_cache}_GRCh38.tar.gz -C /opt/ref/vep-cache/
fi
mamba activate ${pn}.vep
mkdir -p ${result}/${sn}/annotation
vep \
-i ${result}/${sn}/snv/${sn}_filtered.vcf.gz \
-o ${result}/${sn}/annotation/${sn}_filtered_vep.tsv \
--offline \
--cache \
--cache_version ${version_vep_cache} \
--everything \
--dir_cache /opt/ref/vep-cache/ \
--dir_plugins /opt/ref/vep-cache/Plugins \
--species homo_sapiens \
--assembly GRCh38 \
--fasta /opt/ref/${version_reference}/${version_reference}.fasta \
--refseq \
--force_overwrite \
--format vcf \
--tab \
--shift_3prime 1 \
--offline
mamba deactivate
fi11. 使用脚本处理注释结果和过滤vcf结果,输出和室间质评要求格式的数据表格
bash
mamba activate ${pn}.cnvkit
if [ ! -f "${envs}/MatchedSnvVepAnnotationFilter.py" ]; then
aria2c https://raw.githubusercontent.com/doujiangbaozi/sliverworkspace-util/main/somatic/MatchedSnvVepAnnotationFilter.py -d ${envs}/
chmod a+x ${envs}/MatchedSnvVepAnnotationFilter.py
fi
${envs}/MatchedSnvVepAnnotationFilter.py \
-e normal_artifact \
-e germline \
-i strand_bias \
-i clustered_events \
--min-vaf=${snv_vaf} \
--min-tlod=${snv_tlod} \
--min-depth=${snv_depth} \
-v ${result}/${sn}/snv/${sn}_filtered.vcf.gz \
-a ${result}/${sn}/annotation/${sn}_filtered_vep.tsv \
-o ${result}/${sn}/annotation/${sn}.result.SNV.tsv
mamba deactivate
12. 使用cnvkit提供工具输出分布图和热图
bash
mamba activate ${pn}.cnvkit
cnvkit.py scatter ${result}/${sn}/cnv/${sn}_tumor_marked.cnr \
-s ${result}/${sn}/cnv/${sn}_tumor_marked.cns \
-i ' ' \
-n ${sn}_normal \
-o ${result}/${sn}/cnv/${sn}_cnv_scatter.png -t &&
cnvkit.py heatmap ${result}/${sn}/cnv/${sn}_tumor_marked.cns \
-o ${result}/${sn}/cnv/${sn}_cnv_heatmap.png
mamba deactivate
13. 使用cnvkit call 根据cnvkit batch输出结果推算拷贝数
bash
mamba activate ${pn}.cnvkit
cnvkit.py call ${result}/${sn}/cnv/${sn}_tumor_marked.cns \
-o ${result}/${sn}/cnv/${sn}_tumor_marked.call.cns
mamba deactivate
14. 编写脚本处理cnvkit输出,计算cnv基因,exon位置,gain/lost,cn数
bash
mamba activate ${pn}.cnvkit
if [ ! -f "${envs}/CnvAnnotationFilter.py" ]; then
aria2c https://raw.githubusercontent.com/doujiangbaozi/sliverworkspace-util/main/somatic/CnvAnnotationFilter.py -d ${envs}/
chmod a+x ${envs}/CnvAnnotationFilter.py
fi
if [ ! -f "/opt/ref/${version_reference}/refGene.txt" ]; then
aria2c -x 16 -s 16 http://hgdownload.cse.ucsc.edu/goldenPath/${version_reference}/database/refGene.txt.gz -d /opt/ref/${version_reference} -o refGene.txt.gz
cd /opt/ref/${version_reference} && gzip -d refGene.txt.gz
fi
python ${envs}/CnvAnnotationFilter.py \
-r /opt/ref/${version_reference}/refGene.txt \
-i ${cnv_min} \
-x ${cnv_max} \
-D ${cnv_dep} \
-f ${result}/${sn}/cnv/${sn}_tumor_marked.call.cns \
-o ${result}/${sn}/cnv/${sn}.result.CNV.tsv
mamba deactivate
15. 编写脚本处理manta的输出,获取最终sv输出结果,起始位置,基因、频率等
bash
mamba activate ${pn}.cnvkit
if [ ! -f "${envs}/SvAnnotationFilter.py" ]; then
aria2c https://raw.githubusercontent.com/doujiangbaozi/sliverworkspace-util/main/somatic/SvAnnotationFilter.py -d ${envs}/
chmod a+x ${envs}/SvAnnotationFilter.py
fi
if [ ! -f "/opt/ref/${version_reference}/refGene.txt" ]; then
aria2c -x 16 -s 16 http://hgdownload.cse.ucsc.edu/goldenPath/${version_reference}/database/refGene.txt.gz -d /opt/ref/${version_reference} -o refGene.txt.gz
cd /opt/ref/${version_reference} && gzip -d refGene.txt.gz
fi
${envs}/SvAnnotationFilter.py \
-r /opt/ref/${version_reference}/refGene.txt \
-s ${sv_score} \
-f ${result}/${sn}/sv/results/variants/somaticSV.vcf.gz \
-o ${result}/${sn}/sv/${sn}.result.SV.tsv
mamba deactivate
16. 根据之前fastp,samtools depth,samtools flagstat,gatk CollectInsertSizeMetrics等输出,给出综合 QC数据
bash
mamba activate ${pn}.cnvkit
if [ ! -f "${envs}/MatchedQcProcessor.py" ]; then
aria2c https://raw.githubusercontent.com/doujiangbaozi/sliverworkspace-util/main/somatic/MatchedQcProcessor.py -d ${envs}/
chmod a+x ${envs}/MatchedQcProcessor.py
fi
${envs}/MatchedQcProcessor.py --bed /opt/ref/${version_reference}/${project_bed} \
--out ${result}/${sn}/stat/${sn}.result.QC.tsv \
--sample-fastp=${result}/${sn}/trimmed/${sn}_tumor_fastp.json \
--sample-depth=${result}/${sn}/stat/${sn}_tumor_marked.depth \
--sample-flagstat=${result}/${sn}/stat/${sn}_tumor_marked.flagstat \
--sample-insertsize=${result}/${sn}/stat/${sn}_tumor_insertsize_metrics.txt \
--normal-fastp=${result}/${sn}/trimmed/${sn}_normal_fastp.json \
--normal-depth=${result}/${sn}/stat/${sn}_normal_marked.depth \
--normal-flagstat=${result}/${sn}/stat/${sn}_normal_marked.flagstat \
--normal-insertsize=${result}/${sn}/stat/${sn}_normal_insertsize_metrics.txt
mamba deactivate
mamba activate ${pn}.fastp
multiqc ${result}/${sn}/ -f -o ${result}/${sn}/qc
mamba deactivate
最终输出
| 文件名 | 备注 |
|---|---|
| report.pdf | 由sliverworkspace设计的图形化分析报告打印输出为PDF |
| 1701/qc/multiqc_report.html | multiqc聚合报告 |
| 1701/1701.result.SNV.tsv | SNV最终突变结果 |
| 1701/1701/cnv/1701_cnv_heatmap.png | CNV结果热图 |
| 1701/cnv/1701_cnv_scatter.png | CNV结果分布图 |
| 1701/cnv/1701.result.CNV.tsv | CNV最终结果 |
| 1701.result.SV.tsv | SV最终结果 |
| 1701.result.QC.tsv | 最终质控结果 |
| 1701/qc/multiqc_report.html | multiqc报告 |
